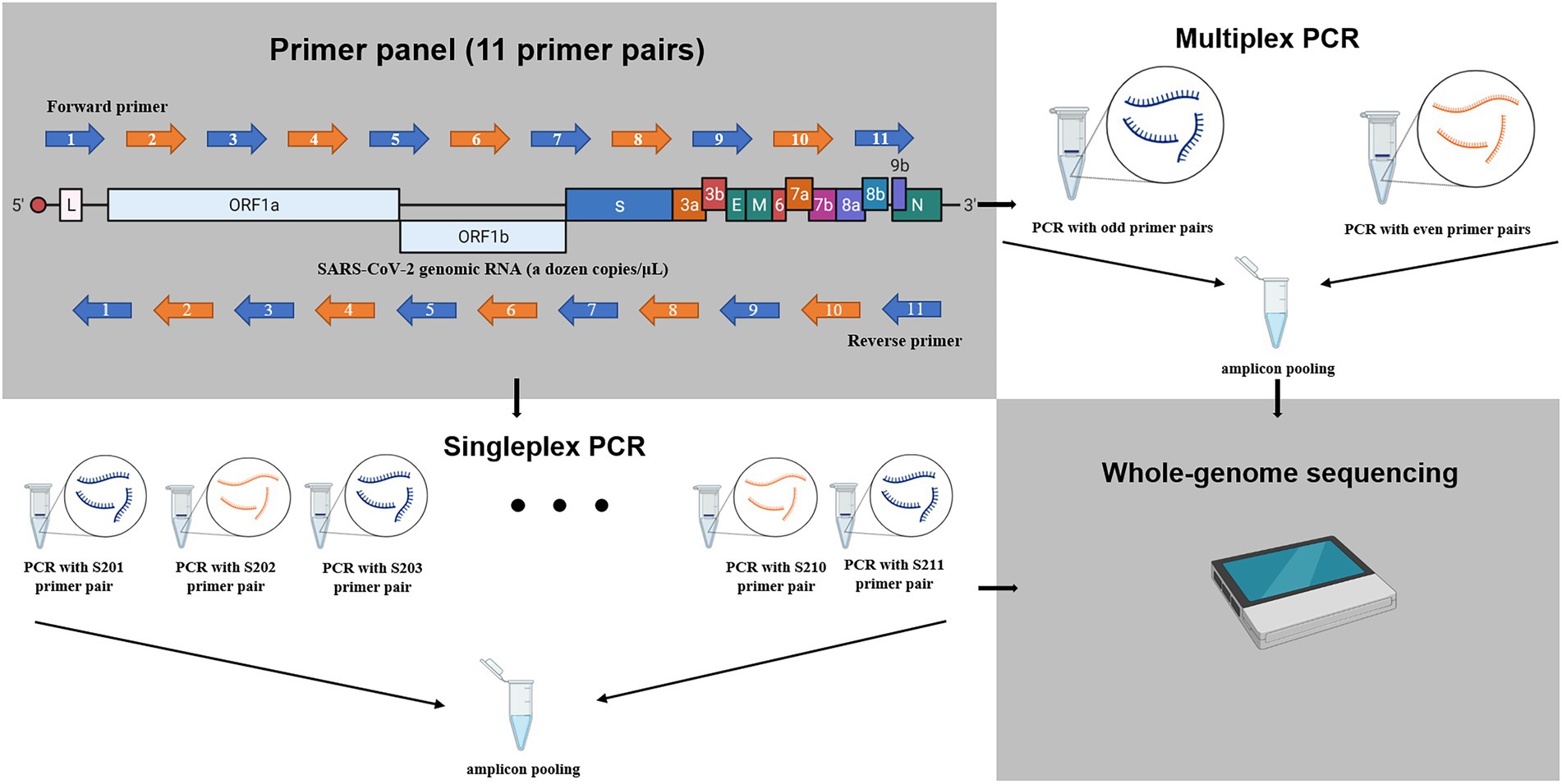
Frontiers | Investigating the Extent of Primer Dropout in SARS-CoV-2 Genome Sequences During the Early Circulation of Delta Variants
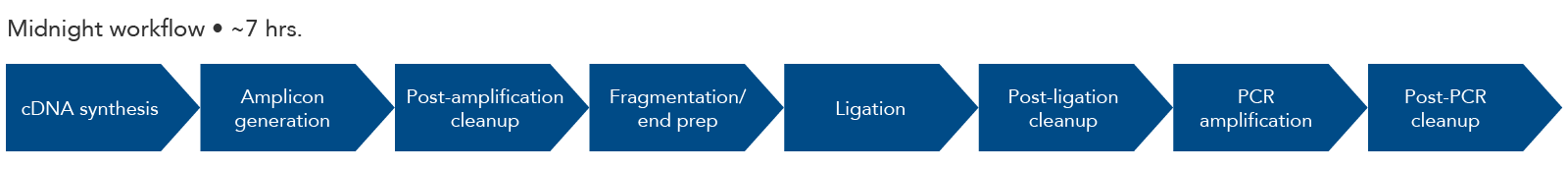
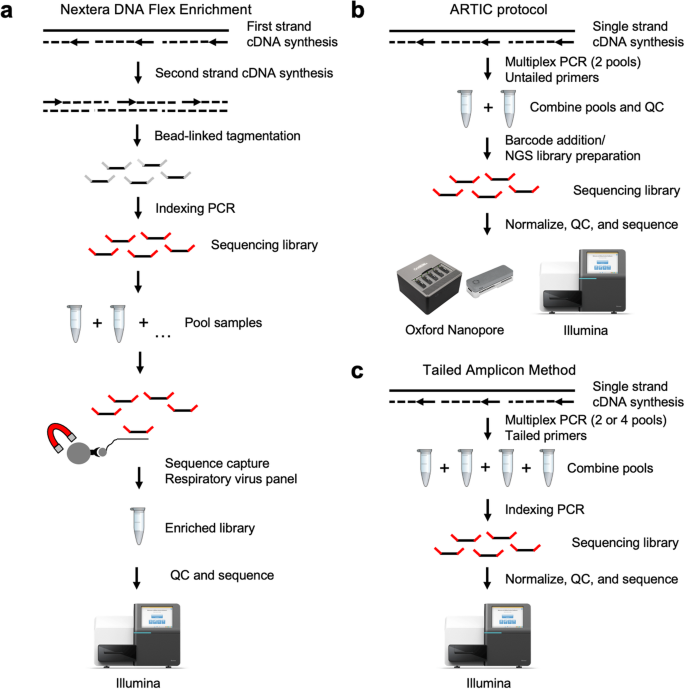
Omicron and beyond: HiFiViral Kit provides labs with a future-proof solution for emerging COVID-19 variants - PacBio
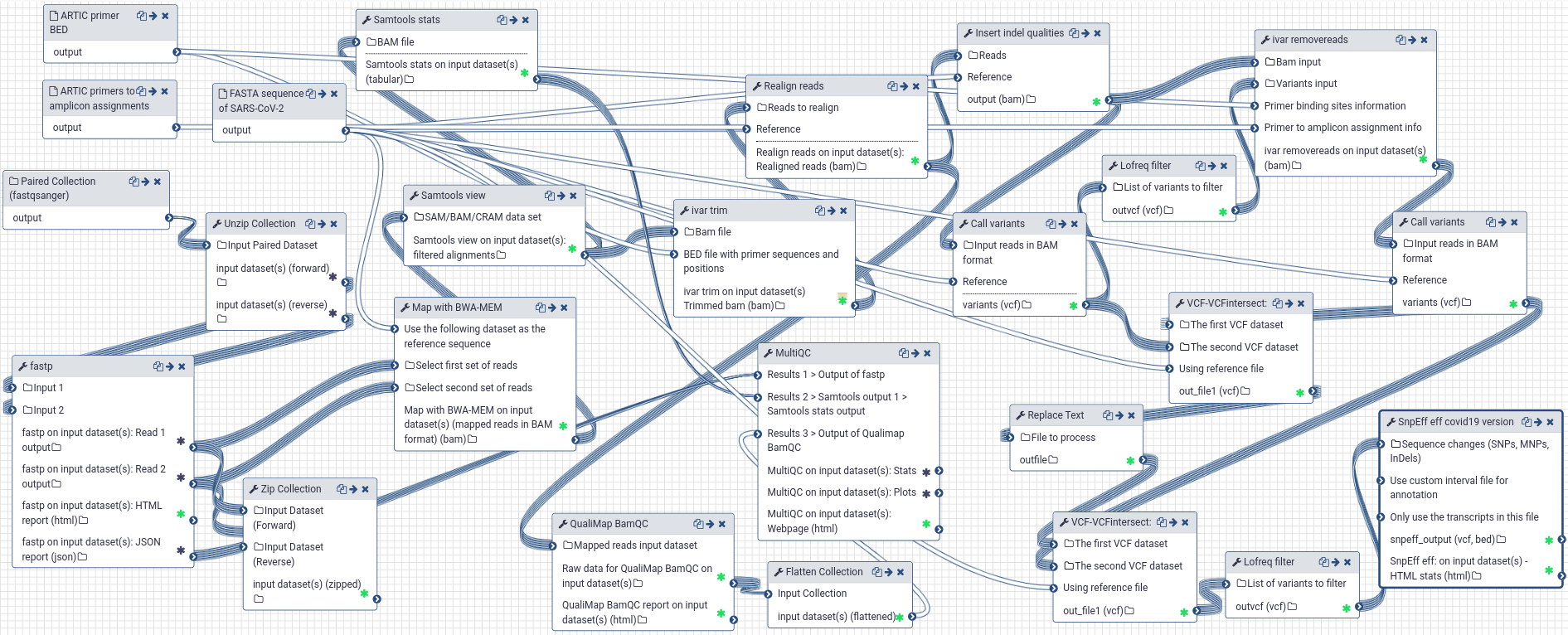
Bioinformatics workflows for SARS-CoV-2; from raw Nanopore reads to consensus genomes using the ARTIC coronavirus protocol

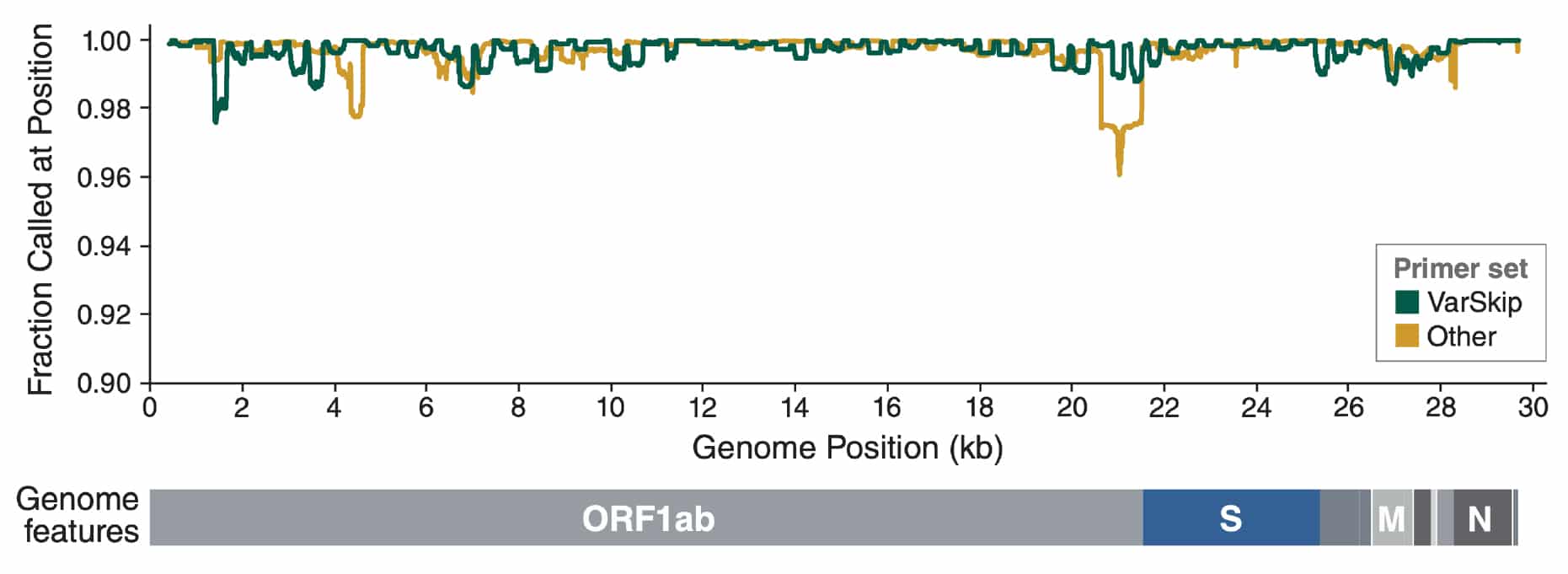
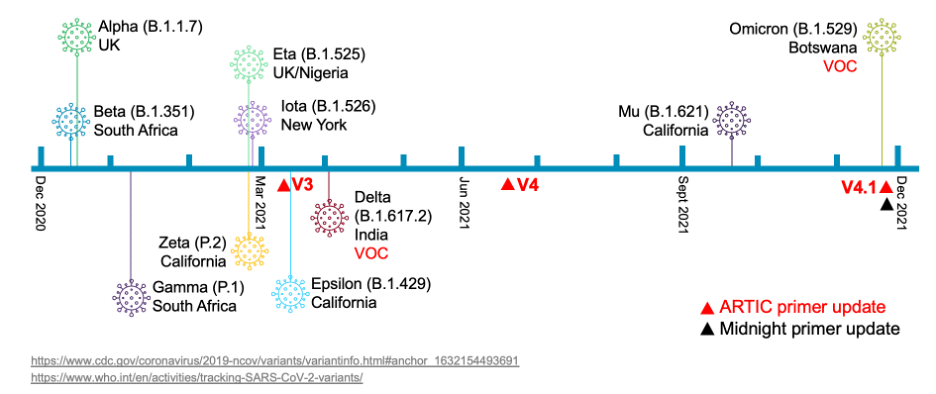
Analysis of the ARTIC Version 3 and Version 4 SARS-CoV-2 Primers and Their Impact on the Detection of the G142D Amino Acid Substitution in the Spike Protein | Microbiology Spectrum
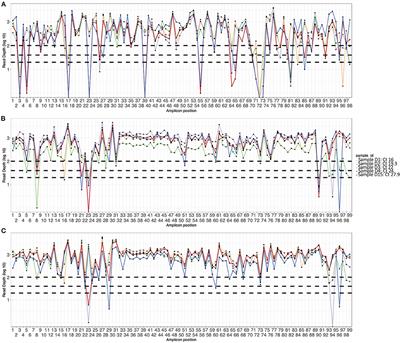
Frontiers | Optimization of the SARS-CoV-2 ARTIC Network V4 Primers and Whole Genome Sequencing Protocol
A proposal of alternative primers for the ARTIC Network's multiplex PCR to improve coverage of SARS-CoV-2 genome sequencing
A Short Plus Long-Amplicon Based Sequencing Approach Improves Genomic Coverage and Variant Detection In the SARS-CoV-2 Genome
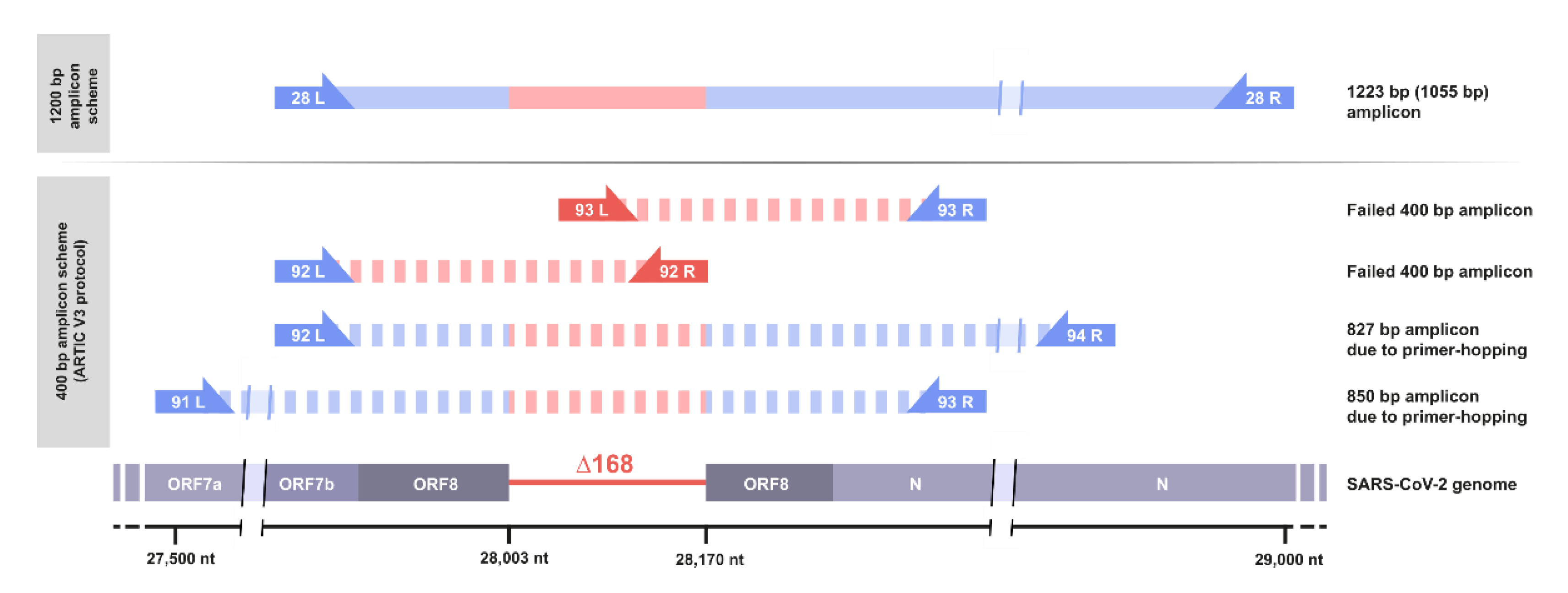
Viruses | Free Full-Text | Multiple Occurrences of a 168-Nucleotide Deletion in SARS-CoV-2 ORF8, Unnoticed by Standard Amplicon Sequencing and Variant Calling Pipelines
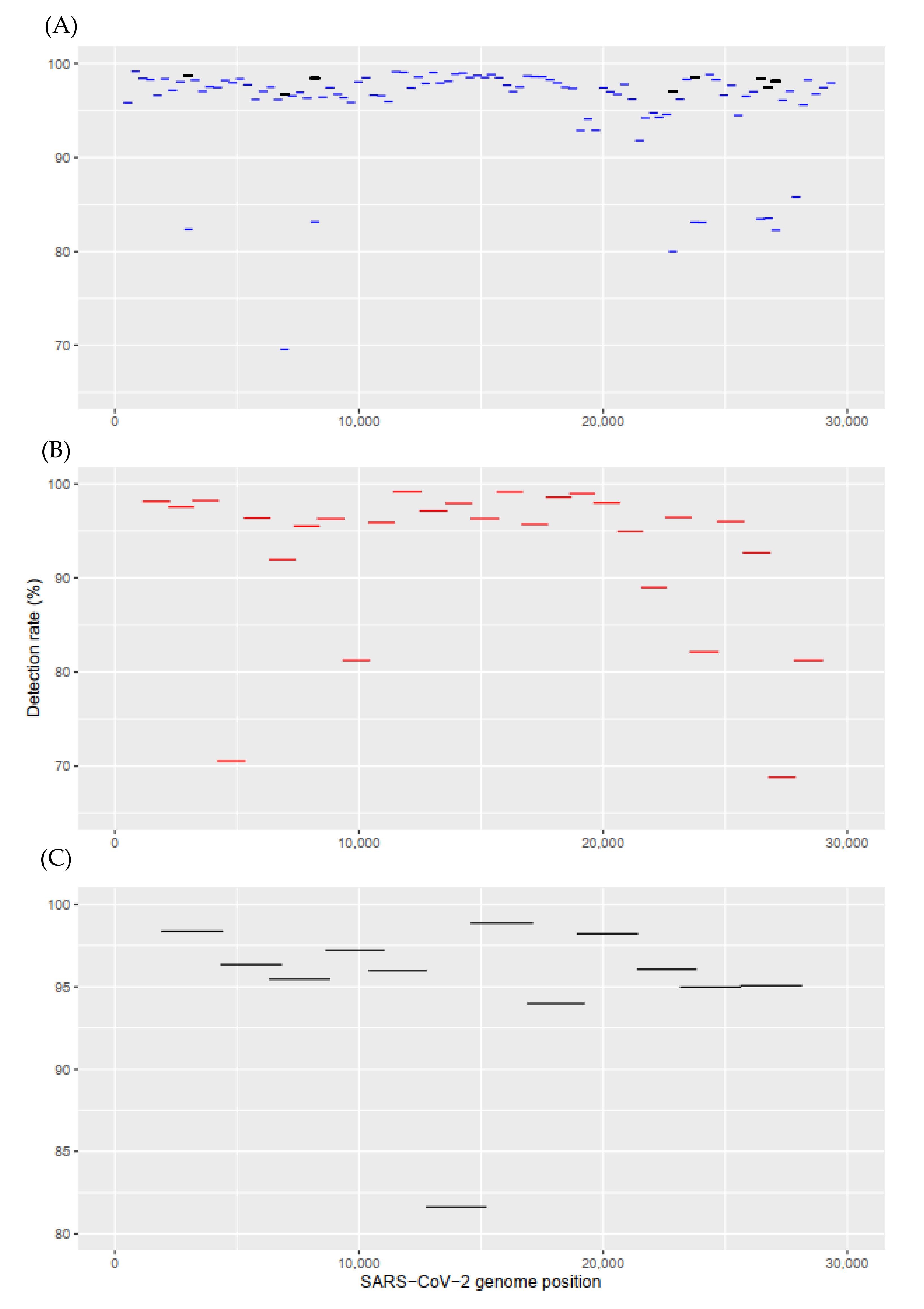
IJMS | Free Full-Text | High-Integrity Sequencing of Spike Gene for SARS-CoV-2 Variant Determination

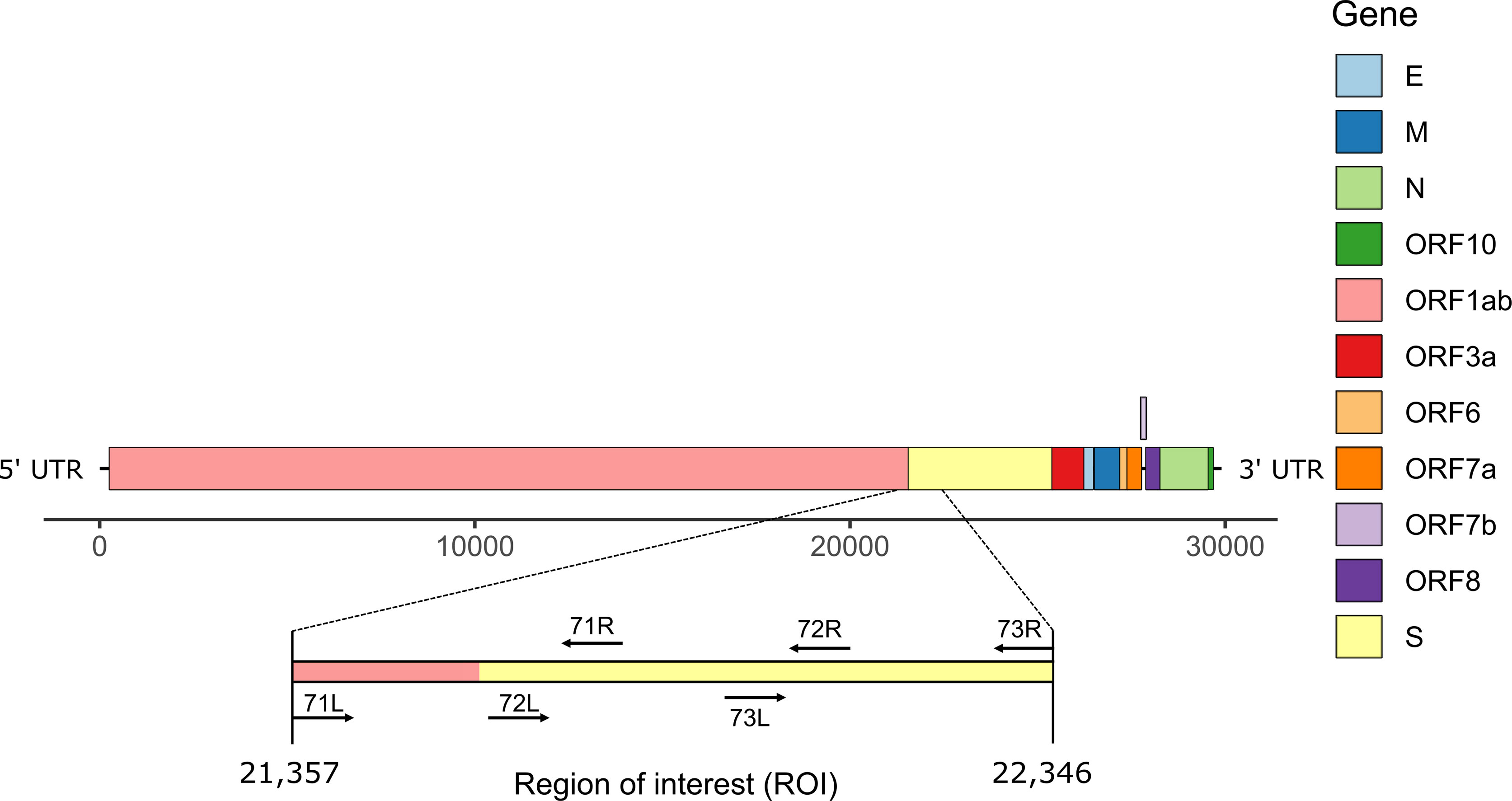
Variation at Spike position 142 in SARS-CoV-2 Delta genomes is a technical artifact caused by dropout of a sequencing amplicon | medRxiv

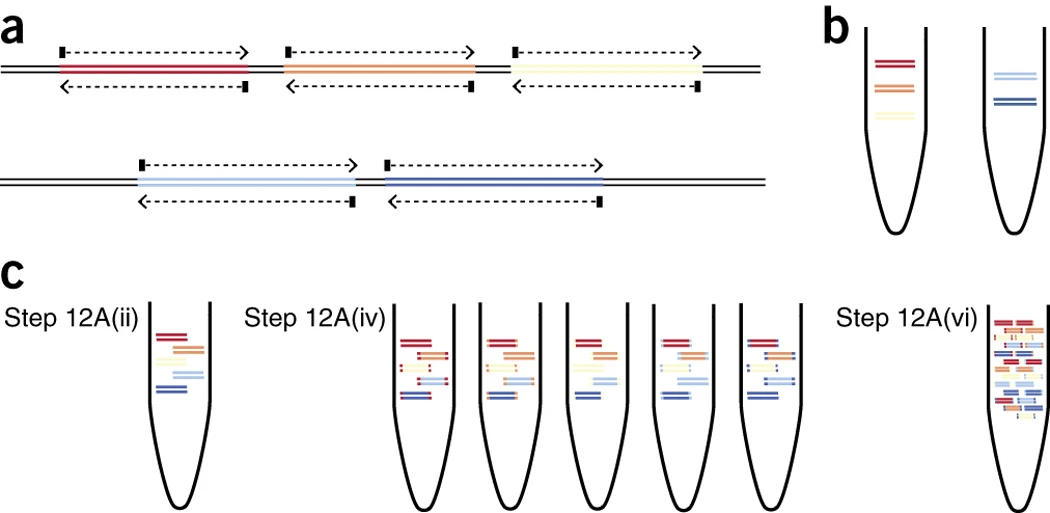
Mini-XT, a miniaturized tagmentation-based protocol for efficient sequencing of SARS-CoV-2 | Journal of Translational Medicine | Full Text

SARS-CoV-2 Genome Sequencing Methods Differ in Their Abilities To Detect Variants from Low-Viral-Load Samples | Journal of Clinical Microbiology